Mapping Genes In Sordaria
Mapping the location of genes
on a chromosome can be accomplished with a diploid organism by following
the percentage of crossover events much like the problems we have done
in class. But what happens when you have a haploid organism like
fungi? In this case, there are no "dominant" or "recessive" traits
(since the organism is haploid). However, the events of recombination
and crossing over can be observed by looking at the spore patterns in the
ascus (spore case of an Ascomycetes). This process is called tetrad
analysis since the outcome of meiosis in Ascomycetes results in a linear
tetrad of haploid spores. By analyzing spore patterns one can observe
crossover events that occurred during meiosis when an allelic marker is
located on each chromatid of a synapsed tetrad. Such an analysis
reveals two important pieces of information about the crossover event.
First, one can determine which two of the four chromatids participated
in the cross over event. Secondly, the gene can be mapped relative
to an observable cytological marker such as the centromere. By determining
the location of several genes, one could eventually determine linkage groups
and chromosome locations for all genes.
Sordaria fimicola
is a common species of ascomycete found on dung. Sordaria
is haploid and spends most of its life cycle in the vegetative state.
Under favorable environmental conditions, different matting types of
Sordaria can undergo sexual reproduction. This occurs by the creation
of the a binucleate hypha, and the eventual fusion of the two nuclei within
a developing ascus fuse to produce a diploid (2n) zygote. This zygote
then undergoes meiosis to form a linear array of haploid ascospores contained
in the ascus (pl., asci). In the case of Sordaria the
meiotic division is followed by a mitotic division to produce eight ascospores
(Figure 1). The asci (about 20) are grouped together within a structure
called the perithecium. It is the dark brown perithecium on the agar plate
that you can observe with the naked eye.

When you observe the Sordaria
in this lab, you will note that the ascospores are of two different colors.
The one most often found in nature is called the wild type (+) and produces
a dark spore. The mutant form of this gene called "tan" (t) produces
a light spore. By observing the order of the ascospores in the ascus
one can determine the order in which the chromosomes are segregated (separated)
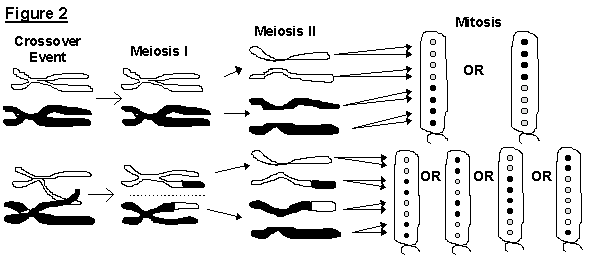
during meiosis. If no crossover events occur, the two genes will
segregate during meiosis I and produced a 4:4 arrangement of ascospores.
If a crossover event does occur, the two genes will not segregate
until meiosis II which will result in a 2:2:2:2 or 2:4:2 sequence of ascospores
(Figure 2).
In the following exercise
you will begin by make a cross between Sordaria with the wild type
ascospore color (dark) and Sordaria with the mutant ascospore color
(tan). Then, after about 11 days of culture, you will observe the
color sequence of ascospores produced in the hybrid asci. From your
observations you will be able to calculate the gene to centromere distance
in map units. Keep in mind that the centromere is telomeric.
PROCEDURE:
Making the Cross:
Each student should obtain
a starch agar petri plate from the instructor. This plate will be
designated the cross plate, and will be used to cross the wild type (dark)
ascospore Sordaria with the mutant type (tan) ascospore Sordaria.
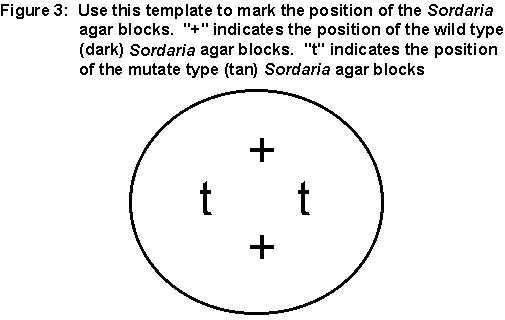
On the bottom of the cross plate write your initials. With the cross
plate upside down, center the petri plate over Figure 3 below. Mark
the "+" and "t" on the bottom of the cross plate. Now turn the plate
right side up. There are two stock plates of Sordaria that
will be circulating round the lab room. One is the wild type (dark)
Sordaria and one is the mutant type (tan) Sordaria.
When the plates come to you, slightly lift the cover of the stock petri
plate and, using a sterile toothpick, transfer a block of the fungi culture
to your starch plate. Place the block upside down over the "+" or
"t" marks, depending on which culture you are using. You will need
two blocks of each type of Sordaria culture.
Leave the cross plate in
the drawer indicated by the instructor. It will take 8 to 10 days
for the perithecia to mature.
Counting Hybrid Asci:
During the laboratory period
when the Sordaria cultures have matured, you will observe
the asci under the compound microscope. Obtain a microscope
slide. Place one drop of water in the center of the microscope slide.
Obtain the Sordaria cross plate that you set-up about a week and
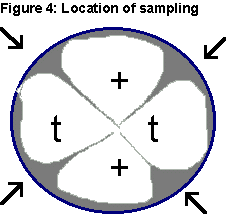
a half ago. Using a toothpick, scrape across the top of the agar
gently to remove several perithecia from the areas designated by arrows
indicated in Figure 4.
 Wash the perithecia off onto
a microscope slide in the drop of water. Obtain a cover slip, place
it over top of the perithecia and gently press the cover slip with your
thumb or a pencil eraser until the perithecia are crushed (you can feel
this as a soft pop). This will release clusters of asci. Start
by using the low power of a microscope, search for hybrid asci (asci that
contain both tan and dark spores) and determine in which area of the cross
plate they were found.
After locating hybrid asci, use high dry magnification to count the number
of Meiosis I (MI) and Meiosis II (MII) asci. Count a total of 200
bi-colored asci, recording the results below. Then calculate the
percent MEIOSIS II and the gene to centromere distance in map units. As
you work, be sure to record the unusual patterns of asci.
Wash the perithecia off onto
a microscope slide in the drop of water. Obtain a cover slip, place
it over top of the perithecia and gently press the cover slip with your
thumb or a pencil eraser until the perithecia are crushed (you can feel
this as a soft pop). This will release clusters of asci. Start
by using the low power of a microscope, search for hybrid asci (asci that
contain both tan and dark spores) and determine in which area of the cross
plate they were found.
After locating hybrid asci, use high dry magnification to count the number
of Meiosis I (MI) and Meiosis II (MII) asci. Count a total of 200
bi-colored asci, recording the results below. Then calculate the
percent MEIOSIS II and the gene to centromere distance in map units. As
you work, be sure to record the unusual patterns of asci.
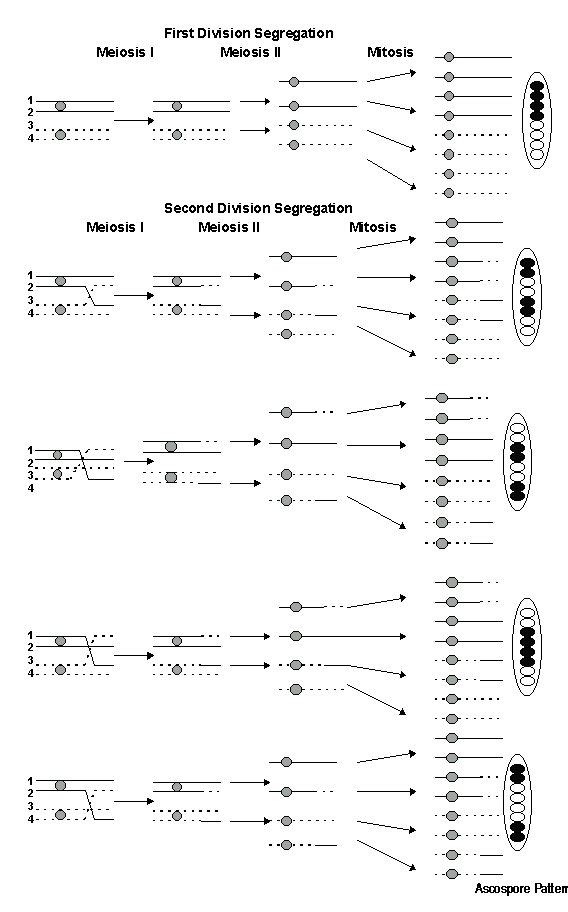
Ascospore Patterns.
Three nuclear divisions are required in the production of an ascus containing
eight ascospores from a diploid zygote. Meiosis I and II result in
the production of the first four haploid nuclei from the diploid zygote.
The eight ascospore nuclei arise from mitosis, the third division.
These three nuclear divisions produce the ascus and the various possible
ascospore patterns of hybrid asci. Segregation of the alleles at
the first meiotic division (MI) results in the sequence 4+:4t or 4t:4+
when no crossing over occurs between the gene and the centromere.
Here the homologous chromosomes segregated during Anaphase I without an
exchange of genetic material between the gene and the centromere.
If crossing over does occur between the gene and the centromere the ascospore
arrangements that follow the second division (MII) segregation will occur.
The 2+:2t:2+:2t arrangement is the result of crossing over between chromatids
two and three, whereas if chromatids one and four crossover, a 2t:2+:2t:2+
pattern results. The 2+:4t:2+ pattern results from crossing over
involving chromatids two and four, whereas crossing over between
chromatids one and three gives the 2t:4+:2t pattern. See Figure 5
below:
Map Distance:
Map distances for haploid organisms can be calculated from the percentage
of crossing over between a gene, in this case gene "t", and the telomeric
centromere. Map distances are estimated on the basis of the frequency
of second division (MII) segregation as follows. First, the total
number of second division (MII) segregation asci are determine. This
number is them divided by the total number of bicolored asci that were
counted. The resulting number is multiplied by 100 to obtain the
percentage of second division segregation. This percentage
of crossovers must be divided in half in order to obtain the map distance
between the gene locus and the centromere. The reason for dividing
the percentage in half deals with the fact that in the second division
segregation asci resulting from a single crossover between the "t" locus
and the centromere only two of the four chromatids actually cross over.
This means that only one half of the possible chromatids are involved.
Thus, half of the chromatids are recombinant and half are parental.
Therefore, the measure of the distance between the gene locus and the centromere
(the map distance) is one half the percentage of second division segregation.
Atypical Segregation Ratios
and Gene Conversion: As you are working with the asci you will
occasionally observe unusual tetrad arrangements. The possible arrangements
are 3:1:1:3, 5:3, and 6:2 which are actually aberrant 4:4 ascospore
patterns. Changes of ascospore genes from wild type to mutant or
mutant to wild type during meiosis can account for these ascospore patterns.
But, the frequency of these type changes are much too great to be
accounted for by spontaneous mutation. Instead, the genes in these
asci are undergoing gene conversion. The asci having
3:1:1:3 or 5:3 ascospore patterns contain two ascospores derived from a
"hybrid" DNA chromatid. Remember that DNA is a complementary double
stranded macromolecule. A hybrid DNA has strands that are noncomplementary
in their nucleotide sequence for the gene in question (i.e. they do not
have the same sequence). Thus when the DNA strands segregate in Mitosis,
the resulting two ascospores end up with different sequences in the genes
affected. The asci ratio 6:2 results from a complete chromatid change,
which could have resulted from a DNA repair system. Studies of aberrant
ascospore patterns and the processes involved in gene conversion have provided
researchers with important information on the molecular mechanisms of recombination.
Read the section in your lecture textbook about the mechanism of gene recombination
and the Holliday model.
Mechanism of Crossing
Over: Two theories have been proposed to describe the involvement
of chiasmata in crossing over. The classical theory maintains that
the chiasmata cause physical strains on the chromosomes and that the location
and presence of the chiasmata area completely random. A chiasmata
may or may not induce breakage and rejoining at the cross over point.
This theory states that the chiasmata are responsible for crossover events
and clearly precede them. Since chiasmata appear during the diplotene
stage of meiosis I, the classical theory holds that the actual crossover
event occurs after diplotene but before the chromosomes separate at anaphase
I. The chiasmatype theory predicts that crossing over precedes chiasma
formation and occurs in pachytene. As a result, in this theory the
chiasmata are formed at points of genetic exchange. Therefore, the
chiasmata seen in the diplotene stage are evidence of crossover events.
Some supporting evidence for the chiasmatype theory comes from the finding
that DNA synthesis occurs in the zygotene stage of meiotic prophase I.
The actual role of this late-replicating DNA is not known. However,
it amounts to 0.3% of the total nuclear DNA and it is distributed randomly
among chromosomes. If this late DNA synthesis is inhibited, chromosomal
synapsis is also inhibited and the process of meiosis is stopped.
This suggests that this DNA synthesis is somehow important for chromosome
alignment during meiosis.
Sordaria Laboratory Worksheet
|
Number of
Meiosis I Asci 4:4
|
Number of
Meiosis II Asci 2:2:2:2 or 2:4:2
|
Total Number of Asci
|
% Meiosis II Asci (#
MII / Total)
|
Gene to Centromere Distance(%
MII / 2)
|
|
|
|
|
|
-
How many different arrangements
did you find? __________________
-
What were the different arrangements?
To answer this question list the black spores as + and the tan (mutant)
spores as t. For example, no crossing over would be recorded as ++++tttt.
-
Which allele is dominant:
the + allele or the t allele? Explain.
-
How do you explain the perithecia
in which the asci contained either eight black or eight tan ascospores?
-
Draw a schematic map of the
telemeric chromosome with the location of the gene.
-
What is the definition of a
"map unit"?
-
How does this procedure, using
the haploid Sordaria, differ from the mapping exercises we have
done with genes on Drosophila chromosomes?
-
What does the aberrant ratio
3:1:1:3 (aberrant 4:4) represent? If you encountered this arrangement,
what frequency did you observe? If you did not encounter this ratio,
what do you know about its frequency?
-
Using drawings like to show
how the 3:1:1:3 arrangement develops in the ascus.
-
How might you account for such
ratios as 5:3 or 3:5? Again diagram the possible crossing pattern
that would give rise to these arrangements.
-
How might you account for such
ratios as 6:2 or 2:6? ? Again diagram the possible crossing pattern
that would give rise to these arrangements.
-
Were any other ascospore arrangements
observed other than the ones previously mentioned? If so, how might
these be explained?
-
By following the incorporation
of tritiated thymidine, it has been demonstrated that a small amount of
DNA synthesis occurs during pachynema. What function might this DNA
synthesis serve? Relate this to what happens in the Holliday (or
Chi) structure.




