Recombinant DNA Technology and Genomics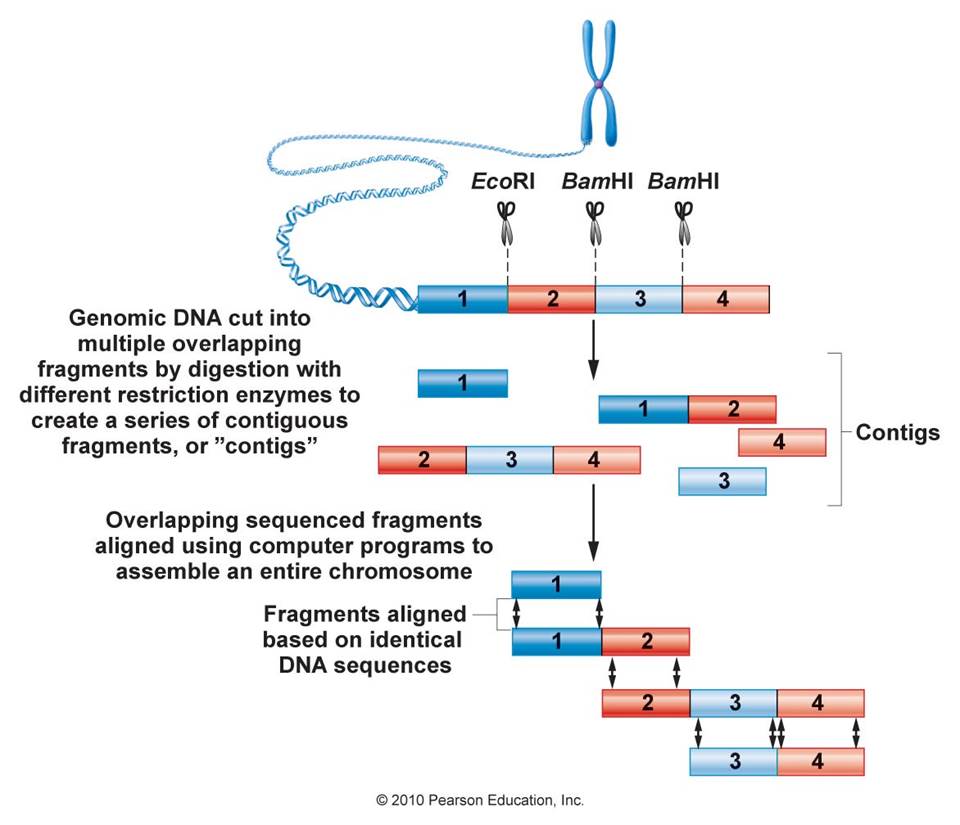
1) List four reasons why we might move DNA from one organism to another.
2) What is a DNA library, and why do you want it to be as small as possible?
3) You can make a library by restriction enzyme digest, pcr, or c-DNA synthesis. How could you use each method to isolate a single gene?
4) If you want to put a eukaryotic gene into a bacterium to see how it works (not just to sequence it), what difficulties do plasmid transfer and restriction enzyme digest methods pose? How could you solve the intron problem?
5) Describe how PCR works and what it is used for.
6) If you do not know anything about the location of a gene but you can isolate the m-RNA, how can you create 1000's of copies of the gene coding sequence?
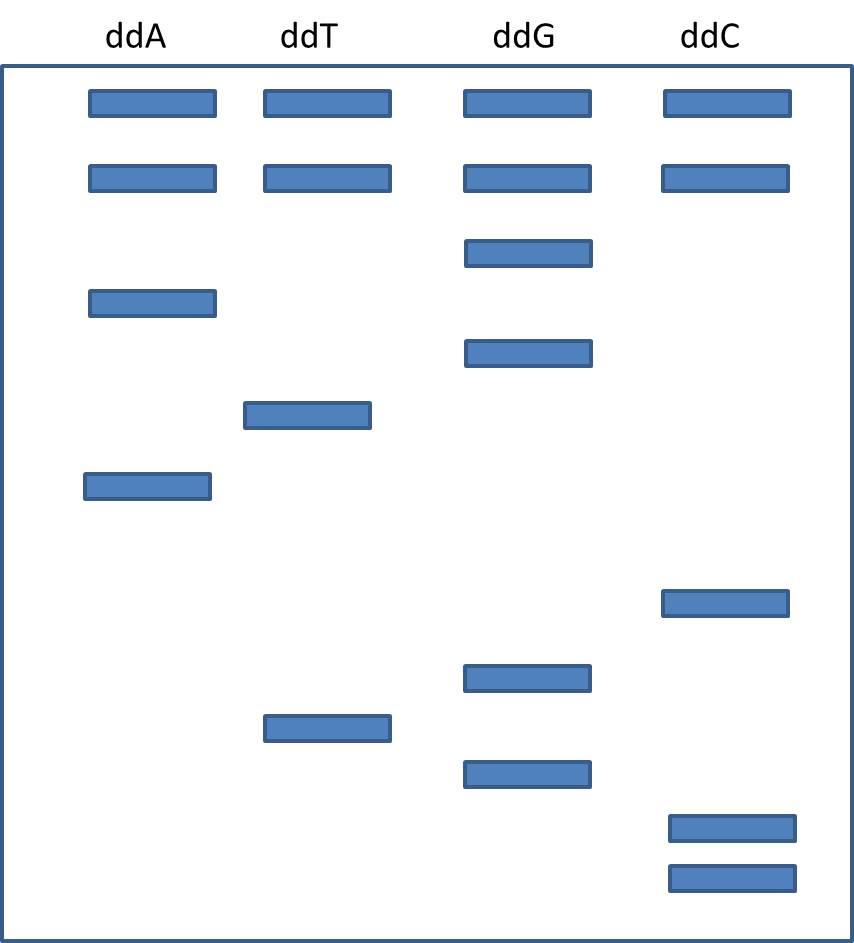
7 ) Describe the logic of adding dideoxy bases to the polymerization reactions to create fragments of different length. Why aren't ALL the adenine triphosphate precursors in the A lane dideoxy bases?
8) Suppose you sequence DNA using the Sanger method, and find the following banding pattern in your gel. What was the sequence?
9) How do you create a "knock-out" mutant and how does this allow us to determine gene/protein function?
10) Two of the most commonly used "genetica modifications" to crops is the insertion of an herbicide resistance gene and the insertion of an "insecticide" gene. Describe how either gene is inserted.
11) How does the overuse of Bt-corn or Bt-cotton in a field affect the evolution of resistance and the long-term utility of these gm plants?
12) What is the basic goal, and methodology, of gene therapy?
13) How does the CRISPR-cas9 system work as a sort of 'immune system' in bacteria?
14) How is it being used for 'gene editing'?